specparam.SpectralTimeModel¶
- class specparam.SpectralTimeModel(*args, **kwargs)[source]¶
Model a spectrogram as a combination of aperiodic and periodic components.
WARNING: frequency and power values inputs must be in linear space. Passing in logged frequencies and/or power spectra is not detected, and will silently produce incorrect results.
- Parameters:
- peak_width_limitstuple of (float, float), optional, default: (0.5, 12.0)
Limits on possible peak width, in Hz, as (lower_bound, upper_bound).
- max_n_peaksint, optional, default: inf
Maximum number of peaks to fit.
- min_peak_heightfloat, optional, default: 0
Absolute threshold for detecting peaks. This threshold is defined in absolute units of the power spectrum (log power).
- peak_thresholdfloat, optional, default: 2.0
Relative threshold for detecting peaks. Threshold is defined in relative units of the power spectrum (standard deviation).
- aperiodic_mode{‘fixed’, ‘knee’} or Mode
Which approach to take for fitting the aperiodic component.
- periodic_mode{‘gaussian’, ‘skewed_gaussian’, ‘cauchy’} or Mode
Which approach to take for fitting the periodic component.
- algorithm{‘spectral_fit’} or Algorithm
The fitting algorithm to use.
- algorithm_settingsdict
Setting for the algorithm.
- metricsMetrics or list of Metric or list or str
Metrics definition(s) to use to evaluate the model.
- convertersdict
Definition for parameter conversions to apply post fitting.
- bandsBands or dict or int or None, optional
Bands object with band definitions, or definition that can be turned into a Bands object.
- debugbool, optional, default: False
Whether to run in debug mode. If in debug, any errors encountered during fitting will raise an error.
- verbosebool, optional, default: True
Verbosity mode. If True, prints out warnings and general status updates.
- **model_kwargs
Additional model fitting related keyword arguments. These are passed into the algorithm object.
Notes
Input power spectra must be provided in linear scale. Internally they are stored in log10 scale, as this is what the model operates upon.
Input power spectra should be smooth, as overly noisy power spectra may lead to bad fits. For example, raw FFT inputs are not appropriate. Where possible and appropriate, use longer time segments for power spectrum calculation to get smoother power spectra, as this will give better model fits.
The time object inherits from the group model, overwriting the data and results objects with versions for fitting models across time. Temporally organized results are collected into the results.time_results attribute, which may include sub-selecting peaks per band (depending on settings). Note that the results.group_results attribute is also available, which maintains the full model results.
- Attributes:
- algorithmAlgorithm
Algorithm object with model fitting settings and procedures.
- modesModes
Modes object with fit mode definitions.
- dataData
Data object with spectral data and metadata.
- resultsResults
Results object with model fit results and metrics.
Methods
__init__(*args, **kwargs)Initialize time model object.
add_data(freqs, power_spectra[, freq_range, ...])Add data (frequencies and power spectrum values) to the current object.
add_modes(aperiodic_mode, periodic_mode)Add modes definition to the object.
convert_results(bands)Convert results to be organized across time.
copy()Return a copy of the current object.
fit([freqs, spectrogram, freq_range, bands, ...])Fit a spectrogram.
get_data([component, space])Get a data component.
get_group(inds[, output_type])Get a new model object with the specified sub-selection of model fits.
get_metrics(category[, measure])Get requested metric(s) from the object.
get_model([ind, regenerate])Get a model fit object for a specified index.
get_params(component[, field])Return model fit parameters for specified feature(s).
load(file_name[, file_path, convert_results])Load time data from file.
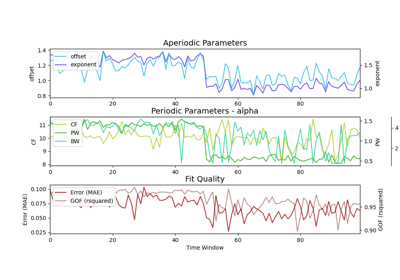
plot([plot_type, save_fig, file_name, file_path])Plot a figure with subplots visualizing the parameters from a SpectralTimeModel object.
print_report_issue([concise])Prints instructions on how to report bugs and/or problematic fits.
print_results([print_type, concise])Print out SpectralTimeModel results.
print_settings([description, concise])Print out the current settings.
report([freqs, spectrogram, freq_range, ...])Fit a spectrogram and display a report, with a plot and printed results.
save(file_name[, file_path, append, ...])Save out results and/or settings from time object.
save_model_report(index, file_name[, ...])"Save out an individual model report for a specified model fit.
save_report(file_name[, file_path, add_settings])Generate and save out a PDF report for models of a spectrogram.
to_df([bands])Convert and extract the model results as a pandas object.
- add_data(freqs, power_spectra, freq_range=None, clear_results=True)¶
Add data (frequencies and power spectrum values) to the current object.
- Parameters:
- freqs1d array
Frequency values for the power spectra, in linear space.
- power_spectra2d array, shape=[n_power_spectra, n_freqs]
Matrix of power values, in linear space.
- freq_rangelist of [float, float], optional
Frequency range to restrict power spectra to. If not provided, keeps the entire range.
- clear_resultsbool, optional, default: True
Whether to clear prior results, if any are present in the object. This should only be set to False if data for the current results are being re-added.
Notes
If called on an object with existing data and/or results these will be cleared by this method call.
- add_modes(aperiodic_mode, periodic_mode)¶
Add modes definition to the object.
- Parameters:
- aperiodic_modeMode or str
Mode for aperiodic component, or string specifying which mode to use.
- periodic_modeMode or str
Mode for periodic component, or string specifying which mode to use.
- convert_results(bands)[source]¶
Convert results to be organized across time.
- Parameters:
- bandsBands or dict or int, optional
How to organize peaks into bands. If Bands or dict, uses band definitions. If int, extracts the first ‘n’ peaks. If not provided, uses band definition available in object.
- copy()¶
Return a copy of the current object.
- fit(freqs=None, spectrogram=None, freq_range=None, bands=None, n_jobs=1, progress=None, prechecks=True, convert_results=True)[source]¶
Fit a spectrogram.
- Parameters:
- freqs1d array, optional
Frequency values for the spectrogram, in linear space.
- spectrogram2d array, shape: [n_freqs, n_time_windows], optional
Spectrogram of power spectrum values, in linear space.
- freq_rangelist of [float, float], optional
Frequency range to fit the model to. If not provided, fits the entire given range.
- bandsBands or dict or int, optional
How to organize peaks into bands. If Bands or dict, uses band definitions. If int, extracts the first ‘n’ peaks.
- n_jobsint, optional, default: 1
Number of jobs to run in parallel. 1 is no parallelization. -1 uses all available cores.
- progress{None, ‘tqdm’, ‘tqdm.notebook’}, optional
Which kind of progress bar to use. If None, no progress bar is used.
- prechecksbool, optional, default: True
Whether to run model fitting pre-checks.
- convert_resultsbool, optional, default: True
Whether to convert results per spectrogram window to be organized over time.
Notes
Data is optional, if data has already been added to the object.
- get_data(component='full', space='log')¶
Get a data component.
- Parameters:
- component{‘full’, ‘aperiodic’, ‘peak’}
- Which data component to return.
‘full’ - full power spectrum ‘aperiodic’ - isolated aperiodic data component ‘peak’ - isolated peak data component
- space{‘log’, ‘linear’}
- Which space to return the data component in.
‘log’ - returns in log10 space. ‘linear’ - returns in linear space.
- Returns:
- output1d array
Specified data component, in specified spacing.
Notes
The ‘space’ parameter doesn’t just define the spacing of the data component values, but rather defines the space of the additive data definition such that power_spectrum = aperiodic_component + peak_component. With space set as ‘log’, this combination holds in log space. With space set as ‘linear’, this combination holds in linear space.
- get_group(inds, output_type='time')[source]¶
Get a new model object with the specified sub-selection of model fits.
- Parameters:
- indsarray_like of int or array_like of bool
Indices to extract from the object.
- output_type{‘time’, ‘group’}, optional
- Type of model object to extract:
‘time’ : SpectralTimeObject ‘group’ : SpectralGroupObject
- Returns:
- outputSpectralTimeModel or SpectralGroupModel
The requested selection of results data loaded into a new model object.
- get_metrics(category, measure=None)¶
Get requested metric(s) from the object.
- Parameters:
- Name of the specific measure(s) to return.
- Returns:
- metricsdict
Dictionary of requested metrics.
- get_model(ind=None, regenerate=True)¶
Get a model fit object for a specified index.
- Parameters:
- indint, optional
The index of the model from group_results to access. If None, return a Model object with initialized settings, with no data or results.
- regeneratebool, optional, default: False
Whether to regenerate the model fits for the requested model.
- Returns:
- modelSpectralModel
The data and fit results loaded into a model object.
- get_params(component, field=None)¶
Return model fit parameters for specified feature(s).
- Parameters:
- fieldstr or int, optional
Column name / index to extract from selected data, if requested. If str, should align with a parameter label for the component fit mode.
- Returns:
- outndarray
Requested data.
- Raises:
- NoModelError
If there are no model fit results available.
- ValueError
If the input for the field input is not understood.
Notes
When extracting peak parameters, an additional column is appended to the returned array, indicating the index that the peak came from.
- load(file_name, file_path=None, convert_results=True)[source]¶
Load time data from file.
- Parameters:
- file_namestr
File to load data from.
- file_pathstr, optional
Path to directory to load from. If None, loads from current directory.
- convert_resultsbool, optional, default: True
Whether to convert results to be organized over time.
- plot(plot_type='time', save_fig=False, file_name=None, file_path=None, **plot_kwargs)[source]¶
Plot a figure with subplots visualizing the parameters from a SpectralTimeModel object.
- Parameters:
- **plot_kwargs
Keyword arguments to apply to the plot.
- Raises:
- NoModelError
If the model object does not have model fit data available to plot.
- static print_report_issue(concise=False)¶
Prints instructions on how to report bugs and/or problematic fits.
- Parameters:
- concisebool, optional, default: False
Whether to print the report in a concise mode, or not.
- print_results(print_type='time', concise=False)[source]¶
Print out SpectralTimeModel results.
- Parameters:
- print_type{‘time’, ‘group’}
Which format to print results out in.
- concisebool, optional, default: False
Whether to print the report in a concise mode, or not.
- print_settings(description=False, concise=False)¶
Print out the current settings.
- Parameters:
- descriptionbool, optional, default: False
Whether to print out a description with current settings.
- concisebool, optional, default: False
Whether to print the report in a concise mode, or not.
- report(freqs=None, spectrogram=None, freq_range=None, bands=None, report_type='time', n_jobs=1, progress=None)[source]¶
Fit a spectrogram and display a report, with a plot and printed results.
- Parameters:
- freqs1d array, optional
Frequency values for the spectrogram, in linear space.
- spectrogram2d array, shape: [n_freqs, n_time_windows], optional
Spectrogram of power spectrum values, in linear space.
- freq_rangelist of [float, float], optional
Frequency range to fit the model to. If not provided, fits the entire given range.
- bandsBands or dict or int, optional
How to organize peaks into bands. If Bands or dict, uses band definitions. If int, extracts the first ‘n’ peaks.
- n_jobsint, optional, default: 1
Number of jobs to run in parallel. 1 is no parallelization. -1 uses all available cores.
- progress{None, ‘tqdm’, ‘tqdm.notebook’}, optional
Which kind of progress bar to use. If None, no progress bar is used.
Notes
Data is optional, if data has already been added to the object.
- save(file_name, file_path=None, append=False, save_results=False, save_settings=False, save_data=False)[source]¶
Save out results and/or settings from time object. Saves out to a JSON file.
- Parameters:
- file_namestr or FileObject
File to save data to.
- file_pathPath or str, optional
Path to directory to load from. If None, saves to current directory.
- appendbool, optional, default: False
Whether to append to an existing file, if available. This option is only valid (and only used) if ‘file_name’ is a str.
- save_resultsbool, optional
Whether to save out model fit results.
- save_settingsbool, optional
Whether to save out settings.
- save_databool, optional
Whether to save out power spectra data.
- save_model_report(index, file_name, file_path=None, add_settings=True, **plot_kwargs)¶
“Save out an individual model report for a specified model fit.
- Parameters:
- indexint
Index of the model fit to save out.
- file_namestr
Name to give the saved out file.
- file_pathPath or str, optional
Path to directory to save to. If None, saves to current directory.
- add_settingsbool, optional, default: True
Whether to add a print out of the model settings to the end of the report.
- plot_kwargskeyword arguments
Keyword arguments to pass into the plot method.
- save_report(file_name, file_path=None, add_settings=True)[source]¶
Generate and save out a PDF report for models of a spectrogram.
- Parameters:
- file_namestr
Name to give the saved out file.
- file_pathstr, optional
Path to directory to save to. If None, saves to current directory.
- add_settingsbool, optional, default: True
Whether to add a print out of the model settings to the end of the report.
- to_df(bands=None)[source]¶
Convert and extract the model results as a pandas object.
- Parameters:
- bandsBands or dict or int, optional
How to organize peaks into bands. If Bands or dict, uses band definitions. If int, extracts the first ‘n’ peaks. If provided, re-extracts peak features; if not, converts from time_results.
- Returns:
- pd.DataFrame
Model results organized into a pandas object.