Note
Go to the end to download the full example code.
Manipulating Objects¶
Examples with combining, sub-selecting, dropping, and averaging power spectrum models.
As you fit power spectrum models, you may end up with multiple model objects, as you fit models within and across subjects, conditions, trials, etc. To help manage and organize the potentially multiple objects that can arise in these cases, here we will explore the utilities offered for managing and organizing within and between model objects.
Using simulated data, in this example we will cover:
combining results across model objects
sub-selecting fits from SpectralGroupModel objects
dropping specified model fits from SpectralGroupModel objects
average across groups of model fits
# Import model object
from specparam import SpectralModel
# Import Bands object, to manage frequency band definitions
from specparam.bands import Bands
# Import utility functions for working with model objects
from specparam.models.utils import average_group, combine_model_objs, compare_model_objs
# Import simulation functions to create our example data
from specparam.sim import sim_power_spectrum
First, we will simulate some example data, and fit some individual power spectrum models.
# Settings for simulations
freq_range = [1, 50]
freq_res = 0.25
# Create some example power spectra
freqs, powers_1 = sim_power_spectrum(\
freq_range, {'fixed' : [0, 1.0]}, {'gaussian' : [10, 0.25, 2]}, nlv=0.00, freq_res=freq_res)
freqs, powers_2 = sim_power_spectrum(\
freq_range, {'fixed' : [0, 1.2]}, {'gaussian' : [9, 0.20, 1.5]}, nlv=0.01, freq_res=freq_res)
freqs, powers_3 = sim_power_spectrum(\
freq_range, {'fixed' : [0, 1.5]}, {'gaussian' : [11, 0.3, 2.5]}, nlv=0.02, freq_res=freq_res)
# Initialize a set of model objects
fm1 = SpectralModel(max_n_peaks=4)
fm2 = SpectralModel(max_n_peaks=4)
fm3 = SpectralModel(max_n_peaks=4)
# Fit power spectrum models
fm1.fit(freqs, powers_1)
fm2.fit(freqs, powers_2)
fm3.fit(freqs, powers_3)
Combining Model Objects¶
Sometimes, when working with models in SpectralModel
or SpectralGroupModel objects, you may want to combine them together,
to check some group properties.
The combine_model_objs() function takes a list of SpectralModel and/or
SpectralGroupModel objects, and combines all available fits together into
a SpectralGroupModel object.
Let’s now combine our individual model fits into a SpectralGroupModel object.
Number of model fits: 3
Note on Manipulating Model Objects¶
Note that these functions that manipulate model objects typically do more than just copy results data - they also check and manage settings and meta-data of objects.
For example, combining SpectralModel objects returns a new SpectralGroupModel object with the same settings.
We can see this by using the compare_model_objs() function to compare
the settings between SpectralModel objects.
You can also use this function if you wish to compare SpectralModel objects to ensure that you are comparing model results that were fit with equivalent settings.
# Compare defined settings across model objects
compare_model_objs([fm1, fg], 'settings')
True
Sub-Select from SpectralGroupModel¶
When you have a SpectralGroupModel object, you may also want to sub-select
a group of models.
Example use cases for this could be:
you want to sub-select models that meet some kind of goodness-of-fit criterion
you want to examine a subset of model reflect, for example, particular channels or trials
To do so, we can use the get_group() method of the
SpectralGroupModel object. This method takes in an input specifying which indices to sub-select,
and returns a new SpectralGroupModel object, containing only the requested model fits.
Note that if you want to sub-select a single model you can
use the get_model() method.
# Define indices of desired sub-selection of model fits
# This could be a the indices for a 'region of interest', for example
inds = [0, 1]
# Sub-select our selection of models from the SpectralGroupModel object
nfg = fg.get_group(inds)
# Check how many models our new SpectralGroupModel object contains
print('Number of model fits: ', len(nfg.results))
Number of model fits: 2
From here, we could continue to do any analyses of interest on our new SpectralGroupModel object, which contains only our models of interest.
Dropping Fits from SpectralGroupModel¶
Another option is to ‘drop’ model fits from a SpectralGroupModel object. You can do this with
the drop() method from a
SpectralGroupModel object.
This can be used, for example, for a quality control step. If you have checked through the object, and noticed some outlier model fits, you may want to exclude them from future analyses.
In this case, we’ll use an example in which we drop any model fits that have particularly high error.
# Drop all model fits above an error threshold
fg.results.drop(fg.get_metrics('error') > 0.01)
Note on Dropped or Failed Fits¶
When models are dropped from SpectralGroupModel objects,
they are set as null models. They are therefore cleared of results, but not literally dropped,
which is done to preserve the ordering of the SpectralGroupModel, so that the n-th model
doesn’t change if some models are dropped.
Note that there may in some cases be Null models in a SpectralGroupModel without explicitly dropping them, if any models failed during the fitting process.
# Check information on null models (dropped models)
print('Number of Null models :\t', fg.results.n_null)
print('Indices of Null models :\t', fg.results.null_inds)
# Despite the dropped model, the total number of models in the object is the same
# This means that the indices are still the same as before dropping models
print('Number of model fits :\t', len(fg.results))
Number of Null models : 1
Indices of Null models : [2]
Number of model fits : 3
FitResults(aperiodic_fit=array([nan, nan]), aperiodic_converted=array([nan, nan]), peak_fit=array([[nan, nan, nan]]), peak_converted=array([[nan, nan, nan]]), metrics={'error_mae': nan, 'gof_rsquared': nan})
Note on Selecting from SpectralModel Objects¶
Both the get_group() and
drop() methods take an input of the indices of the
model(s) to select or drop.
In both cases, the input can be defined in multiple ways, including directly indicating the indices as a list of integers, or boolean masks.
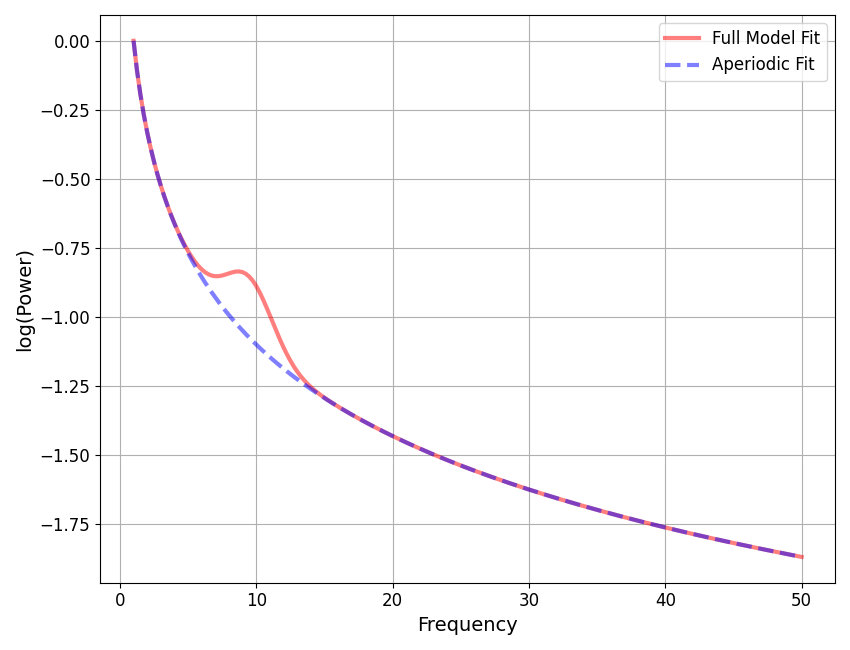
Averaging Across Model Fits¶
Finally, let’s average across the models in our SpectralGroupModel object, to examine the average model of the data.
Note that in order to be able to average across individual models, we need to define a set of frequency bands to average peaks across. Otherwise, there is no clear way to average across all the peaks across all models.
# Define the periodic band regions to use to average across
# Since our simulated data only had alpha peaks, we will only define alpha here
bands = Bands({'alpha': [7, 14]})
# Average across individual models fits, specifying bands and an averaging function
afm = average_group(fg, bands, avg_method='median')
# Plot our average model of the data
afm.plot()
Total running time of the script: (0 minutes 0.284 seconds)